 Figure from"Molecular Biology of the Cell, Fourth Edition"by Alberts et al.
Figure from"Molecular Biology of the Cell, Fourth Edition"by Alberts et al.I've written before about some of the interesting features of intracellular bacteria, but this is possible one of the more exciting and fun things that they do. The picture above shows a eukaryote (human) cell outlined as a green oblong. Within the cell are lots of invasive bacteria (the red dots) some of which have a beautifully long green 'comet tail' flying out behind them.
That comet tail isn't just for show, it is vitally important for movement. The inside of a eukaryote cell is a fairly crowded and busy place, bacteria can't just swim around inside the cell like they would in the wild. Instead they have to rely on physical methods to push them through the cell and like invading virus's (which I wrote about here) they hijack machinery inside the cell to move them around.
Virus particles can latch onto the intracellular transportation machinery to hitch a free ride, but bacteria are too big for that. Instead, what most of them do is to produce proteins known as nucleation-promoting factors. These co-opt cellular proteins (the Arp2/3 complex for anyone with a background in actin polymerisation) which form branched actin fibres behind the bacterial cell, pushing it forward. The 'comet tail' pattern seem above, is seen by using a green stain for the structural actin protein, so you can see it forming long fibrous complexes behind the bacteria. These actin tails can move the bacteria wherever they want to go in the cell, and can also help with the invasion of neighbouring cells. This process is shown diagrammatically below:
That comet tail isn't just for show, it is vitally important for movement. The inside of a eukaryote cell is a fairly crowded and busy place, bacteria can't just swim around inside the cell like they would in the wild. Instead they have to rely on physical methods to push them through the cell and like invading virus's (which I wrote about here) they hijack machinery inside the cell to move them around.
Virus particles can latch onto the intracellular transportation machinery to hitch a free ride, but bacteria are too big for that. Instead, what most of them do is to produce proteins known as nucleation-promoting factors. These co-opt cellular proteins (the Arp2/3 complex for anyone with a background in actin polymerisation) which form branched actin fibres behind the bacterial cell, pushing it forward. The 'comet tail' pattern seem above, is seen by using a green stain for the structural actin protein, so you can see it forming long fibrous complexes behind the bacteria. These actin tails can move the bacteria wherever they want to go in the cell, and can also help with the invasion of neighbouring cells. This process is shown diagrammatically below:

Figure from"Molecular Biology of the Cell, Fourth Edition"by Alberts et al.
The actin system is a good one to use, as actin is ubiquitous inside all eukaryotic cells. In normal cell conditions is it used for structural purposes and is vital in cell division. Also important for the pathogenic bacteria (which after all does not want to kill its host straight away) is that none of these important cellular processes are compromised by the bacteria 'borrowing' some of the actin to move around with.
Another interesting point is that different intracellular bacteria often produce different types of actin tails. L. monocytogenes and S. flexneri have short, highly crosslinked filaments producing short stubby little tails, whereas Rickettsia species have actin tails that are composed of distinctly longer bundles of unbranched actin filaments. Part of the reason for this is that different bacteria will produce different nucleation-promoting factors and some of the more lazy ones (i.e S. flexneri) don't even bother to do that and just use the host nucleation-promoting factors within the invaded cell! Recent work has shown that Rickettsia on the other hand, doesn't even rely on the host Arp2/3 complex to polymerase the actin and instead relies almost entirely on their own, bacterial, proteins.
They truly are beautiful to look at though. Even without all the fancy colour staining:
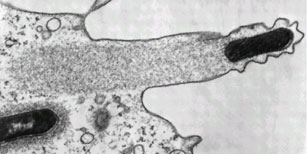
Listeria monocytogenes pushing right at the cell membrane, with actin tail behind. Electron micrograph picture taken from the se reference below
Another interesting point is that different intracellular bacteria often produce different types of actin tails. L. monocytogenes and S. flexneri have short, highly crosslinked filaments producing short stubby little tails, whereas Rickettsia species have actin tails that are composed of distinctly longer bundles of unbranched actin filaments. Part of the reason for this is that different bacteria will produce different nucleation-promoting factors and some of the more lazy ones (i.e S. flexneri) don't even bother to do that and just use the host nucleation-promoting factors within the invaded cell! Recent work has shown that Rickettsia on the other hand, doesn't even rely on the host Arp2/3 complex to polymerase the actin and instead relies almost entirely on their own, bacterial, proteins.
They truly are beautiful to look at though. Even without all the fancy colour staining:

Listeria monocytogenes pushing right at the cell membrane, with actin tail behind. Electron micrograph picture taken from the se reference below
---
Ray K, Marteyn B, Sansonetti PJ, & Tang CM (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nature reviews. Microbiology, 7 (5), 333-40 PMID: 19369949
Kuo SC, & McGrath JL (2000). Steps and fluctuations of Listeria monocytogenes during actin-based motility. Nature, 407 (6807), 1026-9 PMID: 11069185
---
Follow me on Twitter!
Ray K, Marteyn B, Sansonetti PJ, & Tang CM (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nature reviews. Microbiology, 7 (5), 333-40 PMID: 19369949
Kuo SC, & McGrath JL (2000). Steps and fluctuations of Listeria monocytogenes during actin-based motility. Nature, 407 (6807), 1026-9 PMID: 11069185
---
Follow me on Twitter!









