Root of a legume, showing the bumpy nodules where the bacteria live
Studies of how bacteria evolve from free-living organisms into mutualistic partners of eukaryotic (i.e not bacterial) cells has been made easier with the availability of large numbers of sequenced genomes. Intracellular bacteria in general tend to have a smaller genome than free-living bacteria as they have fewer changing environmental conditions to respond too, and many of their metabolic needs can be met by their host. Examining bacteria within their natural environment (rather than the very specific and controlled laboratory environment) also helps to identify the selective forces that can act on bacteria to drive them towards adaptations for mutualistic living.
In order to adapt to a mutualistic lifestyle the bacteria must also gain new genes, which produce proteins that allow it to invade, and communicate with, its host. Genes involved in host interactions are often found in genomic islands, or near to mobile elements which are able to move the genes between organisms (particularly bacteriophages – virus’s that invade bacteria). It has been suggested that the ability of these mobile elements to leave one bacteria and invade neighbouring ones can help to prevent bacterial ‘cheaters’ i.e those that reap the benefits of the mutualistic relationship while relying on the surrounding bacteria to produce the necessary proteins.
Although several species have been found that are thought to have moved from being mutual partners to free-living, the reduction in genome size and host dependency means that once a relationship with the host is established, it tends to remain (occasionally breaking down into parasitism). In fact, once becoming so established that the host carried part (or all) of the bacterial genome, mutualist relationships can often evolve into organellar relationships, where the bacteria become part of the surrounding host cell, and is unable to survive on its own. Established mutualistic bacteria often lose the mobile elements that helped them establish in the first place, and the genome begins to break down as unnecessary genes are slowly lost.
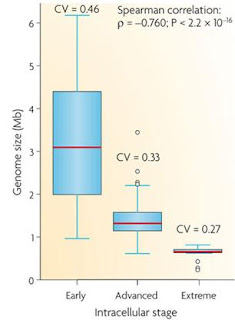
Correlation between genome size and the stage of adaptation to an intracellular lifestyle
Although genome size reduction is the overall fate of bacteria that form a partnership with eukaryotic cells, the process is more than simply the slow loss of genes over time. Different evolutionary pressures will act on both bacteria and host organisms at different stages in the process of creating a relationship which in the early stages requires the gain of genes allowing host interaction. In later stages, the bacterium will lose genes, in some cases passing them on to the safer environment of the host nucleus, which in turn can lead to the bacteria losing all its personal autonomy and becoming a eukaryotic organelle.
---
Toft C, & Andersson SG (2010). Evolutionary microbial genomics: insights into bacterial host adaptation. Nature reviews. Genetics, 11 (7), 465-475 PMID: 20517341