There are several reasons for this. The most important one is that the paycheques for my science-project have just stopped coming. However I do want to actually finish the damn thing, so I've taken up a part time job in a library to get me some money to survive while I'm doing that. I'll also be doing tuitions, supervisions and some exciting new bloggy stuff (can't say what it is right now but Watch This Space) just to scrape together a bit more cash. This means my day will have to be juggled between three hours of actual paid work, doing Science, organising lesson plans, marking essays, planning (and writing) blog posts and doing housework. None of those things can really be left un-done for more than a day.
Pages
New Year
There are several reasons for this. The most important one is that the paycheques for my science-project have just stopped coming. However I do want to actually finish the damn thing, so I've taken up a part time job in a library to get me some money to survive while I'm doing that. I'll also be doing tuitions, supervisions and some exciting new bloggy stuff (can't say what it is right now but Watch This Space) just to scrape together a bit more cash. This means my day will have to be juggled between three hours of actual paid work, doing Science, organising lesson plans, marking essays, planning (and writing) blog posts and doing housework. None of those things can really be left un-done for more than a day.
Levels of evolution
It's helped by the fact that bacterial genomes aren't completely random, and can usually be seperated into 'core' genes and 'accessory' genes. Core genes are most useful to taxonomists as they show what the bacterial species actually is, and where it sits within molecular phylogeny. The accessory genes are more interesting to bacteriologists as the play a more significant role in phenotypic differences and determin what the bacteria do. Paradoxically, the accessory genes are also the ones used most in adaption (think of antibiotic resistance) and are therefore more likely to be evolutionarily selected for or against. These accessory genes are often found in specific 'hypervariable' regions of the genome called genomic islands.
 Cartoon by Nick Kim, www.nearingzero.net
Cartoon by Nick Kim, www.nearingzero.netAs with most new microbiology technologies, these new techniques are initially being applied to human pathogens, as human pathogen research is (rightly) where most of the money is. Using wholescale sequencing technologies on pathogens such as Clostridium difficile which has high levels of antibiotic resistance. The grand aim is to try and understand both the extent and the distribution of natural genomic variation between one bacterial species. This could help to understand what roles are played by bacterial migrations, recombinations (switching DNA around), active selection and drift in the spread of antibiotic resistance.
New Look!
Guest post: Communication - it's not just for cells!
This guest post comes from a real life friend of mine (yes I do have friends in real life). He's currently doing a PhD in developmental biology, and has just started an awesome new blog.
Genetic modification! Stem cells! Genomes! Cloning! These “buzz” words are constantly used by the media in discussions of biological research. These are the words that incite panic and fear in the average tabloid reader. The phrases that make politicians cut science funding, and that make people run screaming when you innocently tell them you work in *deep, ominous, scary voice* science.
Hi, I’m Ret, a good friend of the Lab Rat. From the real world! I also spend far too much of my time locked away in labs, although I work on slightly bigger systems than bacteria. I am currently studying for a PhD in developmental biology. I also strongly believe in the power of good publicity. I believe that scientists need to work much harder to clearly and accurately report their findings to the general public.
For hundreds of years science has been the fodder of the academic, providing all the great advancements as it filters down from the ivory towers of learning, but to the person on the street it is the stuff of horror films; creating patched together monsters and unspeakable abominations of nature. In recent years, with unlimited access to the Internet and the increasing dominance of the media over public opinions, people have really started to take an interest in what we’ve been doing all of these years. An interest that has been fuelled by the economic downturn; the people want to know what’s been happening to all their money. There was a time when we would have rejoiced that people are finally paying attention, yet instead many of us use the typical academic response; we hide behind long words, inaccessible terminology and confounding acronyms.
Having just graduated from university I am very aware of the emphasis that many courses now place on the importance of good presentation skills, which enable us to stand up at conferences and tell a room full of our peers all about what we’ve been doing and why it is significant. What we are not currently taught is how best to communicate with those who are less fluent in the ways of academic research; How to simplify and “sex up” our findings so that people from any background can easily grasp and understand the benefits of what we are doing.
Up until now we have been leaving this job to the media. In the world of journalism, however, a high readership is generally more important than complete scientific accuracy. The eyecatching headlines that result from this are at the root of many public misconceptions of biosciences and only by committing ourselves to direct communication with as many different groups of people as possible can we hope to rectify these grave and long standing errors.
A current favourite of mine is the recent NASA announcement of bacteria which substitute phosphorus in their cells for arsenic, thus providing a new basis for life; or so they claim, even going so far as to imply that this is strong proof for the existence of aliens, and is a revolution in our understanding of science. This is a classic example of the misrepresentation of biological research, (although, having read the paper, it sounds like the research in itself may be a little dodgy). It is however a great example of how to drum up good publicity. I discuss this in more detail on my own blog, but as a starting point, it seems rather odd to me that NASA happens to make this big announcement about discovering potential proof of aliens just days after once again receiving a lot of bad press for once again delaying the shuttle launch. Can anyone say distraction technique?
If we do not engage with the populace, if we do not help them see why our work is important, then they will continue to shy away from us. Without public support all fields of science are subject to unnecessary limitations. Stem cell research globally was significantly inhibited by the short-lived US research ban, a policy brought about by public fear. Terror is keeping the human race from making the most of what it has, we are unable to alleviate poverty and starvation because we are prevented from widespread use of GM crops. Our work with animals is so misunderstood that many who work on animal models are forced to suffer constant hounding by protesters. Most researchers in our fields are here because they want to help; they want to cure the sick, feed the starving and ultimately save the world. However, to see the benefits of this we must bring about acceptance of our ideas so that they can be widely implemented where they are most needed. Hence, communication of our research should be as much a part of our lives as doing the experiments in the first place.
A major factor in this miscommunication, that we all need to work hard to address, is the conception of time. In the 21st century, everyone is used to getting what they want right now! Unfortunately that’s just not the way science works. Start talking about oncogenes and people expect the cure for cancer in the next 6 months. Mention pluripotency and before long the media will be raving about regrowing organs. Influenza, HIV etc. They want it now. Why haven’t we done this? Why haven’t we done that?
The idea that people get from the media, and even from science education up to late degree level is that science is quick. When you did practicals in science class you mixed some stuff together and got a really clear answer in an hour or two. It was perfect, definitive, clean and quick. What you don’t realise until you start doing your own research out at the boundaries is that it’s all a fix. Many modern techniques, even the simple ones involve extensive preparation and incubation times. You can easily spend a week on one set of samples just for them all to fail to work out and for you to get no results. Real significant research is often a very slow process involving many repeated experiments and huge amounts of failures and this is something that many other people just don’t understand; something which is truly foreign to the journalists reporting our findings and to the majority of their readers and something which is easily glossed over, even in our own writings. As it stands, it is only when you do research that you come to appreciate just how much effort went into every tiny detail. How every gene, every protein, every interaction is the product of dozens of PhD projects.
It is for us, as the new generation of academics and science professionals to go against the status quo, to make our work as accessible as possible to everyone and to actively engage with people on a level that they can relate to. Only by doing this can we hope for people to see the true potential benefits of the work we dedicate our lives to, and to understand that there is no quick and easy shortcut to the answers that we are seeking. Only through this can we hope to restore faith in research and save our field from a fast approaching demise at the hands of economic collapse.
The focus of my blog will be accessible science; I hope you recommend it to all of those friends and relations who are always asking you about the latest science news stories. I also invite you to write a guest post, I am looking to cover a broad range of different fields and would be extremely grateful if you felt like sharing your current research with the rest of the world. I also aim to accurately report on the big bioscience news stories. So if you think there is a big story that I should be covering; a serious case of distorting the facts, please let me know and I’ll do my best to provide reliable coverage.
Guest post - Microbes and Madness
Microbes and Madness
A broad range of pathogens are known to cause psychiatric sequelae, including worms (neurocysticercosis), protozoa (cerebral malaria, toxoplasmosis), viruses (HIV, herpes simplex encephalitis, rabies), prions (Creutzfeldt-Jakob disease, kuru), and, of course, bacteria (neurosyphilis, Lyme disease, post-streptococcal syndromes). However, in the spirit of this blog, this post will be focusing on bacteria.
There are essentially four mechanisms through which bacteria cause psychiatric symptoms in humans:
II. Bacteria can trigger a powerful systemic inflammatory response that results in a disruption in brain function.
III. Bacteria can trigger an adaptive immune response which produces antibodies that cross-react with host central nervous system proteins.
IV. Bacteria can be the objects of a phobia.
The range of possible psychiatric presentations is vast. Syphilis, in particular, can mimic any psychiatric syndrome, and was a common diagnosis in psychiatric inpatients a century ago. The possible range of presentations include delirium, dementia, psychosis, mania, and personality changes. Lesions of the frontal lobes are associated with personality changes and disinhibited behaviour, whereas those of the temporal and parietal lobes are associated with cognitive decline. Lyme disease can also mimic several different psychiatric syndromes, but typically affects the limbic system, causing disorders of emotional regulation, including panic attacks, phobias, depression, and obsessive-compulsive behaviour.
The second mechanism listed refers to sepsis-associated delirium. No human organ system is a closed system, including the central nervous system. Bacterial infections with a focus outside the outside the brain are capable of causing a systemic reaction, which affects the brain. The result is an acute confusional state, or delirium.
Common causes are pneumonias and urinary tract infections, although infections of other organ systems are also frequently implicated. Delirium presents as a transient global disorder of cognition. Typically, there is clouding of awareness, disorientation, impaired attention, fluctuating alertness with agitation or drowsiness, hallucinations, illusion, and vague delusions. The state is thought to be caused by a global disruption of brain function, which may result from the effects of a systemic inflammatory response to infection. These effects may include systemic vasodilation causing cerebral hypoperfusion, increased permeability of capillaries allowing toxins to cross the blood-brain barrier, the action of inflammatory cytokines on the brain, and increased body temperature resulting in an increase in neuronal oxygen demand.
The third mechanism is seen following infections with group A beta-haemolytic Streptococcus pyogenes, such as scarlet fever and tonsillitis. In response to infection, the adaptive immune system produces antibodies against antigens on the invading pathogen. However, some streptococcal antigens are similar in some way to antigens on host tissues, and so the antibodies produced mistakenly recognise and attack the host tissues. Examples of post-streptococcal autoimmune diseases include rheumatic fever, glomerulnephritis, and Sydenham’s chorea.
A psychiatric syndrome caused by this mechanism is PANDAS, which stands for paediatric autoimmune neuropsychiatric disorder associated with streptococcus. This typically presents as a dramatic onset of obsessive-compulsive disorder, tic disorders, or Gilles de la Tourette syndrome following an infection with group A beta-haemolytic Streptococcus pyogenes in childhood. It is thought to be a result of autoimmune damage to the basal ganglia, which is the part of the brain involved with the initiation and regulation of motor commands. Interestingly, it has also been suggested that encephalitis lethargica, a mysterious syndrome which caused an epidemic during World War I, may also be caused by a post-streptococcal autoimmune reaction.
The fourth and final mechanism listed refers to mysophobia, or the pathological fear of germs. Behavioural symptoms include repeated washing of hands, excessive cleanliness, and avoidance of social contact. Anxiety and panic attacks also occur. Although the behavioural manifestations are similar, mysophobia is not to be confused with obsessive-compulsive disorder. The former is a phobic disorder, in which the fear of germs underlies the behaviour, and the function of the behaviour is avoidance of the phobic object. In the latter, the behaviour is compulsively carried out in response to the obsession that the behaviour must be carried out.
I hope to have provided an comprehensive overview of some of the interesting ways microbes can cause mental and behavioural disturbances in humans. The function of this ability is open to speculation. The film 28 Days Later tells the story of an artificial ‘Rage’ virus. When a human is infected, he or she becomes uncontrollably aggressive, attacking other humans and infecting them with viruses in the process. Thus, the viruses’ effect on human behaviour is clearly advantageous to their spread and propagation. However, outside of fiction, the advantages of pathogens’ effects on human behaviour is less obvious. Even with rabies, on which the symptoms of the ‘Rage’ were based, there has been no documented human-to-human transmission through bites. In fact, the only documented cases of human-to-human transmission of rabies were of transplant recipients receiving corneas from infected donors! It is therefore not known what evolutionary advantage, if any, the psychiatric sequelae of infection convey to the pathogens. It is possible that they are epiphenomenal.
---
Pfister D, Siegemund M, Dell-Kuster S, Smielewski P, Rüegg S, Strebel SP, Marsch SC, Pargger H, & Steiner LA (2008). Cerebral perfusion in sepsis-associated delirium. Critical care (London, England), 12 (3) PMID: 18457586
Neurosyphilis: Considerations For A Psychiatrist Mark A. Ritchie, Joseph A. Perdigao, Mark A. Ritchie
The Neuropsychiatric Assessment of Lyme Disease
The MolBio Carnival is here!
 The fifth issue of the MolBio carnival is here! We've got loads of great entries in this edition, all focusing on the mysterious world inside cells, so take some time out to take a look and comment on them. The inside of cells is such a fascinating place to explore - and many of these posts were written by the people whose job it is to explore them. I've used a fairly broad interpretation of molecular biology, so in this carnival you'll see everything from the atomic details of protein interactions, to the (comparatively) far bigger world of bacterial colonies in the gut.
The fifth issue of the MolBio carnival is here! We've got loads of great entries in this edition, all focusing on the mysterious world inside cells, so take some time out to take a look and comment on them. The inside of cells is such a fascinating place to explore - and many of these posts were written by the people whose job it is to explore them. I've used a fairly broad interpretation of molecular biology, so in this carnival you'll see everything from the atomic details of protein interactions, to the (comparatively) far bigger world of bacterial colonies in the gut.All good explorations should start with a map - and you don't get much better than the truly gorgeous pictures spotlighted by E. Campbell of the HighMag Blog. This beautiful picture shows a cell with the actin-binding proteins stained purple in order to see how they interact with a mutant actin motor.
Once we head inside the cell, we can start to explore the many complex and fascinating interactions that help to control it. While the DNA might encode all the information needed to create cellular proteins, it isn't just the DNA that is responsible for cellular behaviour, as explained by Christopher Dieni in "How I Learned to Stop Worrying and Love Epigenetics" (which comes second place in the Lab Rat award for best post name). As well as proteins, DNA expression is also controlled by fragments of RNA, explained beautifully clearly by student blogger Khalil A. Cassimally who looks into whether miRNA might be used to control cocaine addiction. And while we're at the level of molecular interactions for cellular control, we can look at control mechanisms for protein folding as well, as the Computational Biology blog takes us through the consequences of entanglement during protein folding.

These small and focused intramolecular reactions aren't just used to control the cell, but also to control far bigger systems, or cell-cell interactions and communication. Memoirs of a Defective Brain explains how the bacteria Strep pyogenes uses intramolecular interactions to prevent the immune system recognising an infection. His post "The SpyCEP who cleaved me" not only wins the Lab Rat award for best post name, but also features the BEST diagram I've ever seen for explaining the subtle and complex interactions between cells of the immune system:

While we're on the theme of bacteria (yay!) we'll head over to the stomach. James, of (currently...) Disease of the Week, has written a great two part series on those bacteria in our gut, focusing on the question of how they actually get into our gut, and what they do when they get there. Part 1 deals with babies, and Part 2 with adults. There's also a lovely post from Lucas Brouwers, of Thoughtomics, which looks at the evolution of cyanobacterial toxins - and why a bacteria that lived millions of years before humans were even thought of would need to produce such a powerful neurotoxin.
And lets not forget the plants! They rely on intracellular interactions as much as any other organism. There's an old (but very good) post from Denim and Tweed about how nitrogen fixing bacteria made the leap from being intracellular parasites to mutualistic helpers. We've also got a post from It Takes 30 - about how sex is specified in plants. Unlike humans, who rely on chromosomes, hormones, and a whole host of social norms and pressures to distinguish the sexes, plants might need no more than a single amino acid insertion.
 Those are basically just brightly painted sexual organs on display
Those are basically just brightly painted sexual organs on display The next edition of the MolBio carnival will be hosted at PHASED, so if you've missed out this time, go submit your posts here by the 3rd of January. Blog carnivals are a great way to share information and to get new readers, so it's highly recommended!
The next edition of the MolBio carnival will be hosted at PHASED, so if you've missed out this time, go submit your posts here by the 3rd of January. Blog carnivals are a great way to share information and to get new readers, so it's highly recommended!Bacterial comet tails
 Figure from"Molecular Biology of the Cell, Fourth Edition"by Alberts et al.
Figure from"Molecular Biology of the Cell, Fourth Edition"by Alberts et al.That comet tail isn't just for show, it is vitally important for movement. The inside of a eukaryote cell is a fairly crowded and busy place, bacteria can't just swim around inside the cell like they would in the wild. Instead they have to rely on physical methods to push them through the cell and like invading virus's (which I wrote about here) they hijack machinery inside the cell to move them around.
Virus particles can latch onto the intracellular transportation machinery to hitch a free ride, but bacteria are too big for that. Instead, what most of them do is to produce proteins known as nucleation-promoting factors. These co-opt cellular proteins (the Arp2/3 complex for anyone with a background in actin polymerisation) which form branched actin fibres behind the bacterial cell, pushing it forward. The 'comet tail' pattern seem above, is seen by using a green stain for the structural actin protein, so you can see it forming long fibrous complexes behind the bacteria. These actin tails can move the bacteria wherever they want to go in the cell, and can also help with the invasion of neighbouring cells. This process is shown diagrammatically below:

Another interesting point is that different intracellular bacteria often produce different types of actin tails. L. monocytogenes and S. flexneri have short, highly crosslinked filaments producing short stubby little tails, whereas Rickettsia species have actin tails that are composed of distinctly longer bundles of unbranched actin filaments. Part of the reason for this is that different bacteria will produce different nucleation-promoting factors and some of the more lazy ones (i.e S. flexneri) don't even bother to do that and just use the host nucleation-promoting factors within the invaded cell! Recent work has shown that Rickettsia on the other hand, doesn't even rely on the host Arp2/3 complex to polymerase the actin and instead relies almost entirely on their own, bacterial, proteins.
They truly are beautiful to look at though. Even without all the fancy colour staining:
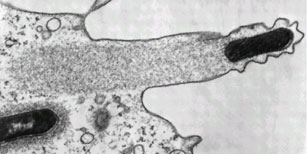
Listeria monocytogenes pushing right at the cell membrane, with actin tail behind. Electron micrograph picture taken from the se reference below
Ray K, Marteyn B, Sansonetti PJ, & Tang CM (2009). Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nature reviews. Microbiology, 7 (5), 333-40 PMID: 19369949
Kuo SC, & McGrath JL (2000). Steps and fluctuations of Listeria monocytogenes during actin-based motility. Nature, 407 (6807), 1026-9 PMID: 11069185
---
Follow me on Twitter!
Oil-eating bacteria

The general rule is that the shorter and fewer rings present, the more toxic the compound to bacteria (ethanol, for example, is deadly) however there are very few hydrocarbons that some bacteria somewhere won't consume. Like all living organisms, bacteria need a carbon source for energy, and when there are none of the coventional ones around (such as glucose and lactose) many of them will start consuming the ones in oil - crude oil contains a lot of carbon, which is why we burn it for energy in the first place.
Oil-eating bacteria have been found in a huge number of locations as well - seawater, freshwater, groundwater, water containing sludge, and land environments such as silt, soil and sand. They are found in arid deserts o the Middle East (which to be fair is a good place to hang out if you consume oil) and even in ice cores from Antarctica. And rather than being found widespread within seperate species, the capacity for oil consupmtion is often found in many different species within a particular location - in the bacterial world you are more likely to share habits with your nearest neighbours than with your closest relatives.
Using these bacteria for our own purposes, however, is proving slightly more problematic. You can purchse freeze-dried hydrocarbon-degrading bacteria and sprinkle them on your oil slick like minature Captain Planets, but their abilities are currently not nearly as effective as chemical detergents, or physical removal of the oil. This is mostly because these freeze-dried bacteria are specialised lab strains that (much like several of the academics that constructed them...) are often highly unsuited to the real world outside of the safe confines of the lab. Sometimes as well the pollutant is unaccessible to the bacteria, which may be floating on the top of the water while the oil is in droplets underneath the surface.
Looking into the potential of oil-eating bacteria is encouraging though, especially after incidents like the BP oil spill earlier this year. For the oil eating bactera that were hanging around, that must have been a feast day! And even though that event has now been largely forgotten by the media, there will still be coils and dropplets of oil floating around from it, and those will be cleaned up almost exclusively by bacteria.
---
Lena Ciric (2010). A natural solution: how bacterial communities can help clean up oil spills Microbiology Today, 229-231
---
Follow me on Twitter!
Breaking Biofilms with DNA
Although living within a biofilm contains significant advantages (protection, good living conditions, etc) there are also times when the bacteria will want to swim away, in order to disperse and form new colonies. The bacteria C. crescentus has an interesting way of doing this, each round of cell division produces two cells: a moving 'swarmer' cell and a non-motile 'stalked' cell which attaches to the biofilm, or any other surface. If conditions are right, the swarmer cells swarmer cells will eventually turn into stalked cells; loosing their flagellum (which are used to swim), retracting their pili, and growing a membranous 'stalk' to attach it to surfaces or surrounding bacteria. This is shown below:

Exactly what it was that turned the motile cells into stable ones and maintained the biofilm was not well understood. Recent research found, rather excitingly, that one factor that could lead to the maintenance of swarmer cells, and the breaking up of biofilms, was extracellular DNA (eDNA) - i.e genetic material that had escaped from cells and was floating around the biofilm. Adding eDNA to C. crescentus biofilms lead to biofilm dispersal, an affect that was reversed by adding DNase enzymes that broke down the DNA.
Why is eDNA such an important signal? Because it's one of the most common products produced from dying cells within a biofilm. Once bacteria in a biofilm die, their cellular integrity breaks down, their insides become their outsides and their genetic material spills out into the surrounding area. This can then act as a powerful signal for surrounding cells, and if the cells around you are dying then where you currently are is clearly not a good place to be. This is also useful when areas of biofilm start to get saturated with too many bacteria, just a few dying off will clear the way for new swarmers to leave and maybe set up colonies elsewhere.
---
Berne C, Kysela DT, & Brun YV (2010). A bacterial extracellular DNA inhibits settling of motile progeny cells within a biofilm. Molecular microbiology PMID: 20598083
Jermy A (2010). eDNA limits biofilm attachment. Nature reviews. Microbiology, 8 (9) PMID: 20737663
---
Follow me on Twitter!
Antibodies
Essential Cell Biology, Second Edition
by Alberts, Bray, Hopkin, Johnson, Lewis, Raff, Roberts, Walter
copyright 2004 by Garland Science Publishing
Storing DNA
Because eukaryotes cells have so much DNA they need to keep it all tightly coiled up, with little molecular tags on the histones to remind them which bits are needed reguarly, and which bits can be mostly ignored. Prokaryotes on the the other hand don't need quite as much control, most of their DNA is going to be used most of the time, but it still needs to be packaged up to fit inside the cell.
The cells do this using by using the unbelieveably eukaryot-ist named "histone-like proteins" such as H-NS which is found in E. coli and related bacteria. Although their precise mechanisms are not quite clear, they have been shown to play a clear role in the coiling of the DNA and maintaining nuceolide stability. They also (like their eukaryote counterpart) help to control gene expression as well as chromatin structure.
In most cases, the H-NS functions as a transcription repressor merely through its physical presence wrapped around DNA - it's harder to transcribe DNA (the first step for protein production) when it's all wrapped up tightly in balls. Some genes though, seem to be activated when associated with H-NS, as it can affect the stability of the mRNA transcript. These are usually genes associated with stress responses; in fact many of the genes that are affected by H-NS are linked to the stress response or changes in the environmental conditions, such as high or low temperature, high osmolarity, changes in pH or oxygen concentration.
Quite how the H-NS controls these parts of the DNA is not entirely clear. Unlike histones, which are positively charged to stabilise the negative charge on the DNA, H-NS proteins are neutral. Although the domain of the molecule that binds to the DNA has been identified, it is not yet certain which parts of these are vital, or how they interact with the DNA molecule. One thing that is clear however is that H-NS are capible of forming dimers, and probably carry out much of their task as a dimer of two joined molecules.
 Domains of an H-NS molecule, from the reference. The oligomerization domain binds to a fellow H-NS while the DNA binding domain attaches to DNA.
Domains of an H-NS molecule, from the reference. The oligomerization domain binds to a fellow H-NS while the DNA binding domain attaches to DNA.Whatever the binding mechanism is, it is unrelated to the actual sequence of the DNA, as H-NS can bind to many different regions of the gene, regardless of sequence. Until recently the H-NS molecules were thought to have no post-translational modifications (unlike histones which are often decorated in molecular markers to indicate which kind of gene they are on) but some have recently been found to be marked with poly-3-hydroxybutyrate, a small lipid molecule. The reason for this is unclear, but it does raise some exciting implications for H-NS control of gene transcription.
When it comes to controling DNA expression, it's clear that in both eukaryotes and prokaryotes, the scaffold proteins that hold the DNA coiled up act as more than just a scaffold. Instead they are involved in a substantial amount of the control of DNA expression, often working closely with other control proteins to ensure the correct genes are turned into proteins.
---
Schröder O, & Wagner R (2002). The bacterial regulatory protein H-NS--a versatile modulator of nucleic acid structures. Biological chemistry, 383 (6), 945-60 PMID: 12222684
---
Follow me on Twitter!
Exciting Things



Antibiotics and gut bacteria
All microbiologists end up writing about gut bacteria at some point. It is the way of things. Disease of the Week is currently doing a whole series on it, and a few weeks ago I covered the interaction of the immune system with gut bacteria (here). However a recent paper came out in Microbiology Today concerning the affect of antibiotics on gut bacteria, which is a topic that I both find interesting and have had some actual experience with.

If you have taken antibiotics it's not worth loosing sleep over, but it's something hospitals are starting to be more and more aware of.
---
Waking sleeping bacteria
Getting into a dormant state is fairly understandable, bacteria have lots of sensing systems which can tell whats going on in their surrounding environment, and dormancy is just a response to that. However getting out of a dormant state is a little more complicated. Once the cell is dormant hardly any of it is active, whatever signal system is used to activate the cells, it has to require very few cellular componants.
In some bacteria (particularly E. coli persisters) exit from dormancy seems to be a fairly random, stochastic effect. Every now and then, one of the cells will simply switch back to being active. If conditions are good it will replicate and re-colonise, whereas if conditions are bad it can either switch back to dormancy or, if it dies, be later replaced as its fellow bacteria come back. It's not a wonderfully good strategy for individuals, but for the species, and for the genes, it works very well.
One possibility the paper in the reference was looking at is that bacteria exit dormancy in response to growth signals from other surrounding bacteria. The idea behind this is that once bacteria start growing and dividing they start secreting signaling molecules such as muropeptides from the cell wall. If these muropeptides are present, it means that bacteria are able to grow and survive, and this might serve as a signal to other bacteria: "conditions are good enough for us!"

With this and the data from stochastic E. coli persisters it is tempting to see this as a full mechanism: exit from dormancy is random, but once one bacteria has grown and survived it can signal to the others that conditions are safe for growth. The only problem with this is that it has the potential to produce large amounts of bacterial growth independent of any signal from the actual environment. It has also been found (via "unpublished observations") that germinating spores don't release the same kind of murapeptide signal, and are unable to activate surrounding spores.
It was also found that the murapeptide signal was not really species specific, which is fascinating from the point of view of bacterial ecology. Growing bacteria, especially Gram-positive species, release large quantities of muropeptides as they grow, and it is a very well conserved molecule, found in the cell wall of other bacteria. A dormant bacteria might therefore not even need one of its own species to report back on conditions outside the cell, it can rely on signals from other bacteria in the surrounding environment.
---
---
Follow me on Twitter!
Interview with the Lab Rat
The more perceptive among you may have noticed that included in the interview is a photo. The photo is of me. After spending two years being paranoid about the internet world connecting a picture of SE Gould (no relation to the Other Gould!) to the persona of Lab Rat, I've gone and sent over a nice big pic of my face for them.
I thought I'd better put forward a few reasons for this.
1 - The ego reason. They wouldn't publish the interview without a picture and I really wasn't about to pass up on an actual interview.
2 - The feminist reason. All the examples they sent me had pictures of men. I really wanted to get a picture of a woman in there somewhere.
3 - The 'I am fed up of being scared and paranoid' reason. This one takes more explanation. Suffice to say because of where I was brought up I was constantly surrounded by this "be doubly careful of everything you do and everyone you talk too" mindset. Even now, back in England, it's everywhere. Walking home in the dark, going travelling alone, being alone in a room with a stranger, I seem to be constantly slightly jumpy that something might hurt me. And all the while I am surrounded by people who are doing the same things as me and don't care about the danger at all. I know what almost all of my bloggy friends look like (with the exception of Psi); they aren't getting mass-murdered so why should I be scared?
But yeah, it was mostly the dazzling prospect of an interview that broke that principle :p
[And anyone who wants to take the picture and feature it in a "top ten sexiest microbiologists!" list: don't. Just don't. It'll piss me off, and will also be a good sign that you need glasses.]
Bacterial cell division and membrane potential

What polylysine does to cells is affect the proton-motive force (pmf), which is used by bacteria to produce energy.
 Diagram shows a stylised version of proton motive force at the bottom of the bacteria. Ions are pumped through the membrane creating energy in the form of ATP. Image taken from Osaka university website.
Diagram shows a stylised version of proton motive force at the bottom of the bacteria. Ions are pumped through the membrane creating energy in the form of ATP. Image taken from Osaka university website.
Cells in the absence (on the left) and the presence (on the right) of CCCP. MinD is attached to GFP, which is bright. Image from the reference.
The moral of the story: Sometimes things don't work for exciting reasons!
(The other moral of the story: always wash your slides :p)
The many ways bacteria move
Most bacteria use their flagella to swim. These are long proteinous threads which look a little like tentacles. Some bacteria just have on of them, while others have most of their body covered in flagella, in order to provide maximum propulsion:
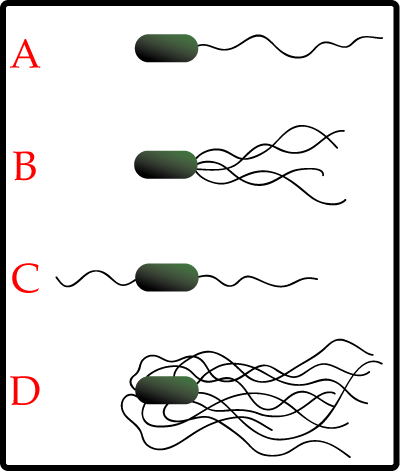
However many bacteria are capible of movement without using flagella, such as the plant and insect pathogen Spiroplasma which moves due to the action of internal filaments. The contractile cytoskeleton is thought to function as a linear motor, meaning the bacteria moves along like a swimmer doing butterfly stroke, through generating a moving kink through the cell, propelling it forward.
Bacteria can also use smaller protrusions called pili, which stick out from the cell surface. There are several different kinds of pili which have different functions (type three are used for pathogenicity, as covered here) and type four pili are commonly used for movement. This is often referred to as 'twitching' as it results in jerky movement. Pili are also used to glide accross surfaces, or even to stand up and walk across them, on little pili legs!
Gliding motility does not always require pili; the bacteria Flavobacterium uses molecules called adhesins to grip to a surface and slide along it. The movement is powered by motors of Gld protein in the bacterial cell wall, as shown in the diagram below (from the reference):

Myococcus Xanthus (which I covered in further detail here) is more likely to use a motor within the cell to create a rotational movement (although movement via polysacharide secretion has also been suggested). The cytoplasmic AglZ protein is thought to act as the motor, as it remains stationary with respect to the surface as the cells glide accross it. This is shown below (image from the reference):

In addition to these active forms of movement, some bacteria also proceed more passively, without using much of their own energy. Aquatic bacteria can use internal gas vesicles in order to rise to the surface (to be closer to sunlight or nutrients). Other bacteria spread simply by growing, pushing bacteria near the surface of the colony into new terratory by all the bacterial growth around it. One of my favourite is the method used by intracellular parasites such as Listeria monocytogenes which polymerises the actin molecules of it's host behind it to travel through the human cell, leaving little superman trails of actin behind it.
There is a huge diversity in the way bacteria move, and interestingly, many of these strategies are also connected to other intracellular processes, after all the mechanism for moving yourself along a surface need not be different to the mechanism for moving molecules around directly under the cell wall. Studying these different ways helps to give an appreciation of the world bacteria live in, and how they organise themselves to survive it.
---
Jarrell KF, & McBride MJ (2008). The surprisingly diverse ways that prokaryotes move. Nature reviews. Microbiology, 6 (6), 466-76 PMID: 18461074
---
Follow me on Twitter!
Chopping bits out of the genome
Streptomyces bacteria, however, have bigger genomes and the luxary to invest in genes which are not strictly necessary for bacterial survival. These are called Secondary metabolite genes (as opposed to the necessary primary metabolites) and they code for genes that form an arsenal of weapons for the Streptomyces to deploy. Most Strep are soil-based, and they need the ability to produce secondary metabolites (such as antibiotics) to fight off invading bacteria, and clear terratory to expand their growth into.
What has recently been done (very ingeniously) is to remove the secondary metabolism genes from the bacterial species Streptomyces avermitilis creating essentially an 'empty' strep bacteria, that can grow and divide but not produce any of the secretory substances that strep are known for. The researchers managed to cut an entire 1.5Mb of DNA right out of the genome - helped by the fact that all the secondary metabolite genes cluster together on one side of the chromosome.
They did this using a common molbio technique, the cre-lox system. "Lox" is an area of DNA and "cre" is the protein that very specifically cuts DNA at the Lox site - it acts like a pair of scissors. They put a Lox site on either end of the DNA that codes for secondary metabolites (using a techinque called recombination in order to attach the lox into the chromosome) along with the DNA for the Cre protein under an inducible promoter. Once the bacteria had grown, they activated Cre production, which then cut the unwanted DNA out of the bacteria. This technique was amazingly successful and is shown diagramatically below (picture from the reference):

The avimitilis now contains no secondary metabolites at all, which makes a wonderful 'empty' system to use for studying how secondary metabolites are made. Genes from other bacteria, or even some of the removed genes, can be added back in, piece at a time, to see how much of the gene is necessary for metabolite production, and which regulatory pathways are the most important. Creating an empty cell also has potential implications for biotechnology, after all when trying to produce antibiotics you want the cell working as hard as possible just to produce your product, not wasting time and resources on other metabolites!
I'm really impressed at this kind of large scale synthetic-biology. It may be an area I end up going into in the near future...
---
Komatsu M, Uchiyama T, Omura S, Cane DE, & Ikeda H (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America, 107 (6), 2646-51 PMID: 20133795

